Bioconductor
Overview
Teaching: XX min
Exercises: XX minQuestions
What is Bioconductor?
How can I use Bioconductor effectively for my analysis?
Objectives
Give an overview of the Bioconductor project including its website
Introduce concepts of reproducibility, coherence, interoperability and stability
Bioconductor
In the previous lesson we have already learned a little bit about the Bioconductor[^Bioconductor] project. In this class we will formalize our understanding of the Bioconductor[^Bioconductor] project.
Important: Remember that Bioconductor packages are downloaded via the function
BiocManager::install().
Bioconductor is a repository that collects open-source software that facilitates rigorous and reproducible analysis of data from current and emerging biological assays in R. In addition, Bioconductor supports development, education and a thriving community. The The broad goals of the Bioconductor project are:
- To provide widespread access to a broad range of powerful statistical and graphical methods for the analysis of genomic data.
- To facilitate the inclusion of biological metadata in the analysis of genomic data, e.g. literature data from PubMed, annotation data from Entrez genes.
- To provide a common software platform that enables the rapid development and deployment of extensible, scalable, and interoperable software.
- To further scientific understanding by producing high-quality documentation and reproducible research.
- To train researchers on computational and statistical methods for the analysis of genomic data.
One of the best ways to explore the Bioconductor project is its website.
The Bioconductor website
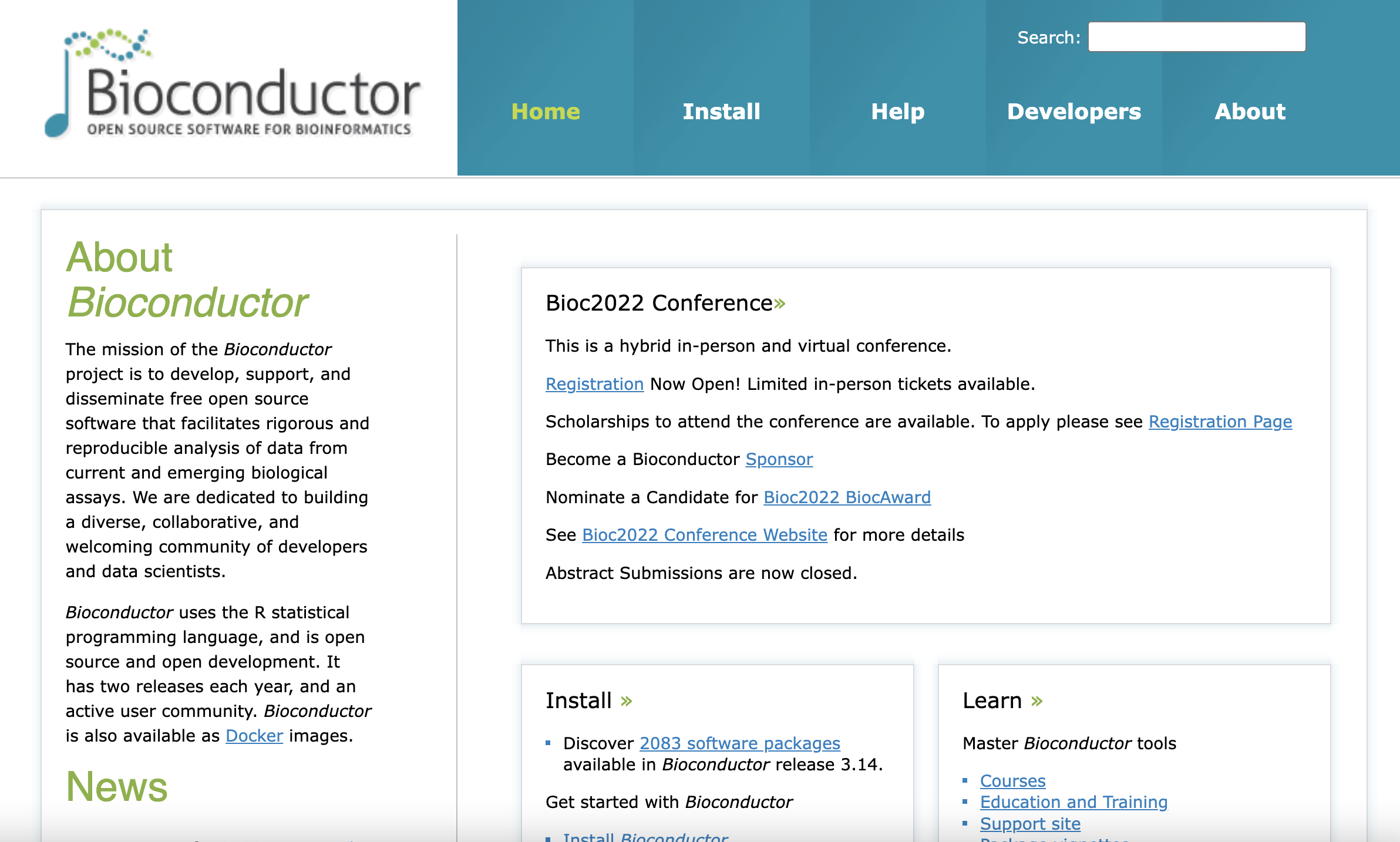
The website tells us that there are over 2,000 packages. This is obviously way too many packages to explore individually. So how would you find potentially useful packages for the analysis of your dataset?
Exploring different topics with biocViews
In Bioconductor, each package is classified as belonging to different categories.
These different categories are called biocViews and they are structured
as follows:
- Software: Packages that provide functions for statistical or graphical methods.
- AnnotationData: Packages that store annotations (i.e genome annotation) and respective access functions.
- ExperimentData: Packages that store example datasets.
- Workflow: Packages that assemble html tutorials using multiple packages for an analysis.
Each category is further divided into additional sub-categories, which are divided into sub-sub-categories that refer to specific assays, techniques, and research fields.
BiocViews can help tremendously with identifying packages that could be of use to you in the analysis of your dataset. These are easily searchable once you have navigated to the page that lists all packages.
Challenge
In your groups, identify a research topic of interest and then use the search function to find packages related to this topic. Think about the following:
- What keywords define your topic of interest?
- What type of packages are good for beginners and why?
- What other information on this website may be useful?
Exploiting the detailed documentation of the Bioconductor project
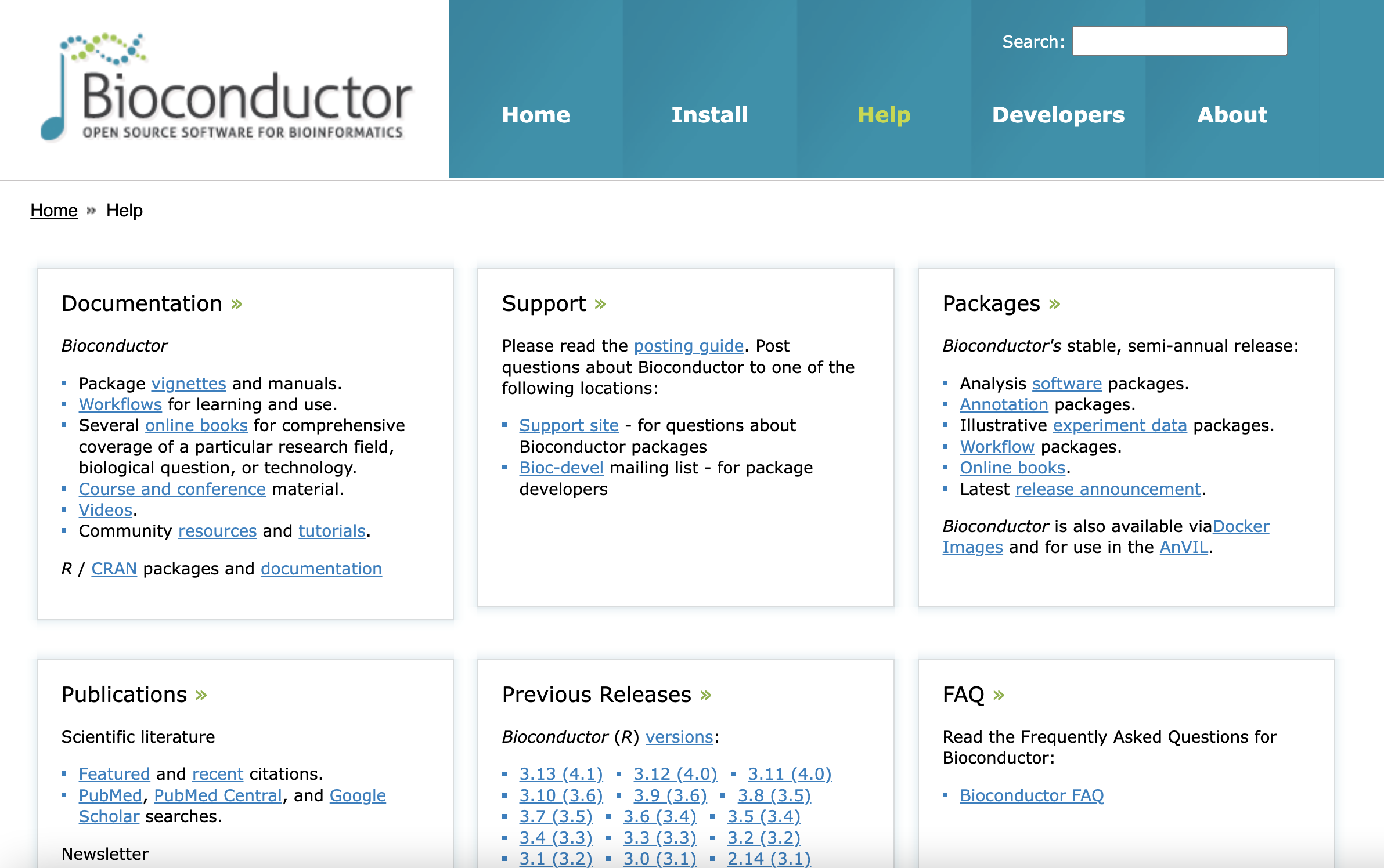
The other part of the Bioconductor website crucial for anyone’s learning journey is the help page. This page collects some outstanding Bioconductor learning resources such as
- Comprehensive books introducing coverage of a research field
- Courses and conference materials
- Videos
- Community resources and tutorials.
- Support site
Most importantly it introduces the concept of vignettes, which are part of
Bioconductor’s mission to enhance reproducibility through rigorous documentation.
In Bioconductor almost every package (certainly every software package) has to
include a vignette. Vignettes are small tutorials that explain common use cases
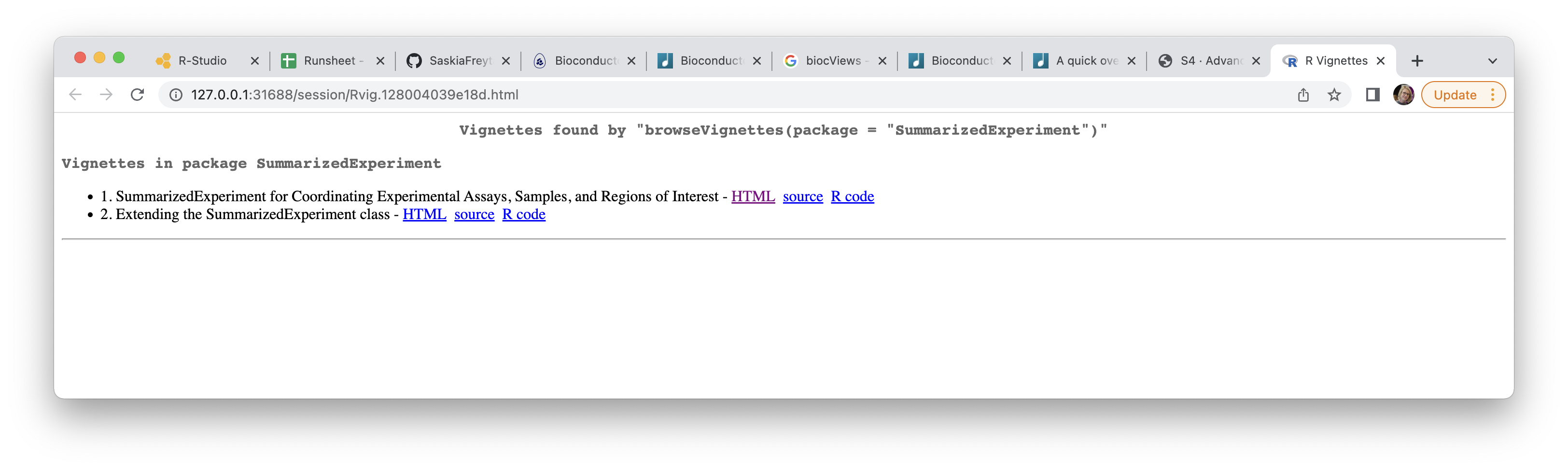
of a package. For example, let’s explore the vignettes available for
SummarizedExperiment:
browseVignettes(package = "SummarizedExperiment")
This should open a separate browser tab that will list availabe vignettes. By
clicking on the html link you will open a nicely formatted tutorial that
you should be easily able to follow. Vignettes are a great place to start when
trying to get familar with a new package.
Core Bioconductor principles
Bioconductor is organized around some core principles:
- interoperablility with existing infrastructure to facilitate reuse and avoid replication
- coherence in coding, documentation and use of existing infrastructure
- rigorous documentation
- reproducibility
- stability ensuring that there are limited clashes due to versions
While it is not absolutely necessary for the end-user to remember these, the core principles explain some of the idiosyncrasies of Bioconductor that you may come across.
S4 classes
Challenge
Use the function
stron theseSummarizedExperiment object you worked with during the last lession. What oddity do you notice? Hint: Compare the output to output ofstrfunction applied torna.Solution
We can see certain elements of the object starting with an @.
S4 programming allows object-oriented programming in R and thus ensure coherence, stability, and interoperability. Object-oriented programming is a computer programming model that organizes software design around data, or objects, rather than functions and logic. An object can be defined as a data field that has unique attributes and behavior.In S4, certain classes are defined with specific accessibility functions (called methods).
# These two statements will result in the same output.
se@colData
DataFrame with 22 rows and 10 columns
title geo_accession organism age sex
<character> <character> <character> <character> <factor>
GSM2545336 CNS_RNA-seq_10C GSM2545336 Mus musculus 8 weeks Female
GSM2545337 CNS_RNA-seq_11C GSM2545337 Mus musculus 8 weeks Female
GSM2545338 CNS_RNA-seq_12C GSM2545338 Mus musculus 8 weeks Female
GSM2545339 CNS_RNA-seq_13C GSM2545339 Mus musculus 8 weeks Female
GSM2545340 CNS_RNA-seq_14C GSM2545340 Mus musculus 8 weeks Male
... ... ... ... ... ...
GSM2545353 CNS_RNA-seq_3C GSM2545353 Mus musculus 8 weeks Female
GSM2545354 CNS_RNA-seq_4C GSM2545354 Mus musculus 8 weeks Male
GSM2545362 CNS_RNA-seq_5C GSM2545362 Mus musculus 8 weeks Female
GSM2545363 CNS_RNA-seq_6C GSM2545363 Mus musculus 8 weeks Male
GSM2545380 CNS_RNA-seq_9C GSM2545380 Mus musculus 8 weeks Female
infection strain time tissue mouse
<factor> <character> <factor> <factor> <factor>
GSM2545336 InfluenzaA C57BL/6 Day8 Cerebellum 14
GSM2545337 NonInfected C57BL/6 Day0 Cerebellum 9
GSM2545338 NonInfected C57BL/6 Day0 Cerebellum 10
GSM2545339 InfluenzaA C57BL/6 Day4 Cerebellum 15
GSM2545340 InfluenzaA C57BL/6 Day4 Cerebellum 18
... ... ... ... ... ...
GSM2545353 NonInfected C57BL/6 Day0 Cerebellum 4
GSM2545354 NonInfected C57BL/6 Day0 Cerebellum 2
GSM2545362 InfluenzaA C57BL/6 Day4 Cerebellum 20
GSM2545363 InfluenzaA C57BL/6 Day4 Cerebellum 12
GSM2545380 InfluenzaA C57BL/6 Day8 Cerebellum 19
colData(se) #colData is a method that allows access to the column meta data
DataFrame with 22 rows and 10 columns
title geo_accession organism age sex
<character> <character> <character> <character> <factor>
GSM2545336 CNS_RNA-seq_10C GSM2545336 Mus musculus 8 weeks Female
GSM2545337 CNS_RNA-seq_11C GSM2545337 Mus musculus 8 weeks Female
GSM2545338 CNS_RNA-seq_12C GSM2545338 Mus musculus 8 weeks Female
GSM2545339 CNS_RNA-seq_13C GSM2545339 Mus musculus 8 weeks Female
GSM2545340 CNS_RNA-seq_14C GSM2545340 Mus musculus 8 weeks Male
... ... ... ... ... ...
GSM2545353 CNS_RNA-seq_3C GSM2545353 Mus musculus 8 weeks Female
GSM2545354 CNS_RNA-seq_4C GSM2545354 Mus musculus 8 weeks Male
GSM2545362 CNS_RNA-seq_5C GSM2545362 Mus musculus 8 weeks Female
GSM2545363 CNS_RNA-seq_6C GSM2545363 Mus musculus 8 weeks Male
GSM2545380 CNS_RNA-seq_9C GSM2545380 Mus musculus 8 weeks Female
infection strain time tissue mouse
<factor> <character> <factor> <factor> <factor>
GSM2545336 InfluenzaA C57BL/6 Day8 Cerebellum 14
GSM2545337 NonInfected C57BL/6 Day0 Cerebellum 9
GSM2545338 NonInfected C57BL/6 Day0 Cerebellum 10
GSM2545339 InfluenzaA C57BL/6 Day4 Cerebellum 15
GSM2545340 InfluenzaA C57BL/6 Day4 Cerebellum 18
... ... ... ... ... ...
GSM2545353 NonInfected C57BL/6 Day0 Cerebellum 4
GSM2545354 NonInfected C57BL/6 Day0 Cerebellum 2
GSM2545362 InfluenzaA C57BL/6 Day4 Cerebellum 20
GSM2545363 InfluenzaA C57BL/6 Day4 Cerebellum 12
GSM2545380 InfluenzaA C57BL/6 Day8 Cerebellum 19
Challenge
Why is it bad practice to use the @ to access parts of an object?
Solution
Using methods functions to access certain parts of the object enhances readibility of your code. If the underlying class structure changes (i.e. the name of the element) the method will still work.
If you want to know more about S4 classes you can find more information in these slides on their implementation in the Bioconductor package.
The release cycle
Bioconductor has two releases each year, typically in April and October, where all packages are updated to their next version in a way that they are interoperable (i.e. they do not clash when you load more than one). The releases conincide with the releases of new R versions, which also happen twice a year. This has two significant implications:
1) To ensure that packages on Bioconductor work flawlessly always use
BiocManager::install to install a package, even when it is not a technically
Bioconductor package. Biconductor mirrors most other R package repositories like
CRAN and so the package will be most likely available. This will avoid clashed
with packages being ahead of the Bioconductor release.
2) You will need to update your Bioconductor packages twice a year (after updating R) to have all the latest versions. Refer to this guide for updating R qnd R-Studio
and this guide for updating Bioconductor.
Working with annotations
Bioconductor provides extensive annotation resources. These can be gene centric, or genome centric. Annotations can be provided in packages curated by Bioconductor, or obtained from web-based resources.
Gene centric AnnotationDbi packages include:
- Organism level: e.g. org.Mm.eg.db.
- Platform level: e.g. hgu133plus2.db, hgu133plus2.probes, hgu133plus2.cdf.
- System-biology level: GO.db Genome centric GenomicFeatures packages include
- Transcriptome level: e.g. TxDb.Hsapiens.UCSC.hg19.knownGene, EnsDb.Hsapiens.v75.
- Generic genome features: Can generate via GenomicFeatures One web-based resource accesses biomart, via the biomaRt package:
- Query web-based ‘biomart’ resource for genes, sequence, SNPs, and etc
The most popular annotation packages have been modified so that they can make use
of a new set of methods to more easily access their contents. These four methods
are named: columns, keytypes, keys and select. They can currently be used with all chip, organism, and TxDb packages along with the popular GO.db package.
An extremely common kind of Annotation package is the so called platform based or
chip based package type. This package is intended to make the manufacturer labels
for a series of probes or probesets to a wide range of gene-based features. A package
of this kind will load an ChipDb object. Below is a set of examples to show how you
might use the standard 4 methods to interact with an object of this type.
First we need to load the package:
suppressPackageStartupMessages({
library(hgu95av2.db)
})
If we list the contents of this package, we can see that one of the many things loaded is an object named after the package “hgu95av2.db”:
ls("package:hgu95av2.db")
[1] "hgu95av2" "hgu95av2_dbconn" "hgu95av2_dbfile"
[4] "hgu95av2_dbInfo" "hgu95av2_dbschema" "hgu95av2.db"
[7] "hgu95av2ACCNUM" "hgu95av2ALIAS2PROBE" "hgu95av2CHR"
[10] "hgu95av2CHRLENGTHS" "hgu95av2CHRLOC" "hgu95av2CHRLOCEND"
[13] "hgu95av2ENSEMBL" "hgu95av2ENSEMBL2PROBE" "hgu95av2ENTREZID"
[16] "hgu95av2ENZYME" "hgu95av2ENZYME2PROBE" "hgu95av2GENENAME"
[19] "hgu95av2GO" "hgu95av2GO2ALLPROBES" "hgu95av2GO2PROBE"
[22] "hgu95av2MAP" "hgu95av2MAPCOUNTS" "hgu95av2OMIM"
[25] "hgu95av2ORGANISM" "hgu95av2ORGPKG" "hgu95av2PATH"
[28] "hgu95av2PATH2PROBE" "hgu95av2PFAM" "hgu95av2PMID"
[31] "hgu95av2PMID2PROBE" "hgu95av2PROSITE" "hgu95av2REFSEQ"
[34] "hgu95av2SYMBOL" "hgu95av2UNIPROT"
We can look at this object to learn more about it:
hgu95av2.db
ChipDb object:
| DBSCHEMAVERSION: 2.1
| Db type: ChipDb
| Supporting package: AnnotationDbi
| DBSCHEMA: HUMANCHIP_DB
| ORGANISM: Homo sapiens
| SPECIES: Human
| MANUFACTURER: Affymetrix
| CHIPNAME: Affymetrix HG_U95Av2 Array
| MANUFACTURERURL: http://www.affymetrix.com
| EGSOURCEDATE: 2021-Apr14
| EGSOURCENAME: Entrez Gene
| EGSOURCEURL: ftp://ftp.ncbi.nlm.nih.gov/gene/DATA
| CENTRALID: ENTREZID
| TAXID: 9606
| GOSOURCENAME: Gene Ontology
| GOSOURCEURL: http://current.geneontology.org/ontology/go-basic.obo
| GOSOURCEDATE: 2021-02-01
| GOEGSOURCEDATE: 2021-Apr14
| GOEGSOURCENAME: Entrez Gene
| GOEGSOURCEURL: ftp://ftp.ncbi.nlm.nih.gov/gene/DATA
| KEGGSOURCENAME: KEGG GENOME
| KEGGSOURCEURL: ftp://ftp.genome.jp/pub/kegg/genomes
| KEGGSOURCEDATE: 2011-Mar15
| GPSOURCENAME: UCSC Genome Bioinformatics (Homo sapiens)
| GPSOURCEURL:
| GPSOURCEDATE: 2021-Feb16
| ENSOURCEDATE: 2021-Feb16
| ENSOURCENAME: Ensembl
| ENSOURCEURL: ftp://ftp.ensembl.org/pub/current_fasta
| UPSOURCENAME: Uniprot
| UPSOURCEURL: http://www.UniProt.org/
| UPSOURCEDATE: Mon Apr 26 21:53:12 2021
Please see: help('select') for usage information
If we want to know what kinds of data are retriveable via select, then we should use the columns method like this:
columns(hgu95av2.db)
[1] "ACCNUM" "ALIAS" "ENSEMBL" "ENSEMBLPROT" "ENSEMBLTRANS"
[6] "ENTREZID" "ENZYME" "EVIDENCE" "EVIDENCEALL" "GENENAME"
[11] "GENETYPE" "GO" "GOALL" "IPI" "MAP"
[16] "OMIM" "ONTOLOGY" "ONTOLOGYALL" "PATH" "PFAM"
[21] "PMID" "PROBEID" "PROSITE" "REFSEQ" "SYMBOL"
[26] "UCSCKG" "UNIPROT"
If we are further curious to know more about those values for columns, we can consult the help pages. Asking about any of these values will pull up a manual page describing the different fields and what they mean.
help("SYMBOL")
If we are curious about what kinds of fields we could potential use as keys to query
the database, we can use the keytypes method. In a perfect world, this method will
return values very similar to what was returned by columns, but in reality, some kinds
of values make poor keys and so this list is often shorter.
keytypes(hgu95av2.db)
[1] "ACCNUM" "ALIAS" "ENSEMBL" "ENSEMBLPROT" "ENSEMBLTRANS"
[6] "ENTREZID" "ENZYME" "EVIDENCE" "EVIDENCEALL" "GENENAME"
[11] "GENETYPE" "GO" "GOALL" "IPI" "MAP"
[16] "OMIM" "ONTOLOGY" "ONTOLOGYALL" "PATH" "PFAM"
[21] "PMID" "PROBEID" "PROSITE" "REFSEQ" "SYMBOL"
[26] "UCSCKG" "UNIPROT"
If we want to extract some sample keys of a particular type, we can use the keys
method.
head(keys(hgu95av2.db, keytype="SYMBOL"))
[1] "A1BG" "A2M" "A2MP1" "NAT1" "NAT2" "NATP"
And finally, if we have some keys, we can use select to extract them. By simply
using appropriate argument values with select we can specify what keys we want to
look up values for (keys), what we want returned back (columns) and the type of
keys that we are passing in (keytype).
#1st get some example keys
k <- head(keys(hgu95av2.db,keytype="PROBEID"))
# then call select
select(hgu95av2.db, keys=k, columns=c("SYMBOL","GENENAME"), keytype="PROBEID")
'select()' returned 1:1 mapping between keys and columns
PROBEID SYMBOL
1 1000_at MAPK3
2 1001_at TIE1
3 1002_f_at CYP2C19
4 1003_s_at CXCR5
5 1004_at CXCR5
6 1005_at DUSP1
GENENAME
1 mitogen-activated protein kinase 3
2 tyrosine kinase with immunoglobulin like and EGF like domains 1
3 cytochrome P450 family 2 subfamily C member 19
4 C-X-C motif chemokine receptor 5
5 C-X-C motif chemokine receptor 5
6 dual specificity phosphatase 1
An organism level package (an ‘org’ package) uses a central gene identifier (e.g.
Entrez Gene id) and contains mappings between this identifier and other kinds of
identifiers (e.g. GenBank or Uniprot accession number, RefSeq id, etc.). The name
of an org package is always of the form org.
Challenge
Display the OrgDb object for the
org.Hs.eg.dbpackage. Use the columns method to discover which sorts of annotations can be extracted from it. Finally, use the keys method to extract UNIPROT identifiers and then pass those keys in to the select method in such a way that you extract the gene symbol and KEGG pathway information for each. Use the help system as needed to learn which values to pass in to columns in order to achieve this.Solution
library(org.Hs.eg.db) uniKeys <- head(keys(org.Hs.eg.db, keytype="UNIPROT")) cols <- c("SYMBOL", "PATH") select(org.Hs.eg.db, keys=uniKeys, columns=cols, keytype="UNIPROT")
A TxDb package (a ’TxDb’ package) connects a set of genomic coordinates to various transcript oriented features. The package can also contain Identifiers to features such as genes and transcripts, and the internal schema describes the relationships between these different elements. All TxDb containing packages follow a specific naming scheme that tells where the data came from as well as which build of the genome it comes from.
Challenge
Display the TxDb object for the
TxDb.Hsapiens.UCSC.hg19.knownGenepackage. As before, use the columns and keytypes methods to discover which sorts of annotations can be extracted from it. Use thekeysmethod to extract just a few gene identifiers and then pass those keys in to theselectmethod in such a way that you extract the transcript ids and transcript starts for each.Solution
library(TxDb.Hsapiens.UCSC.hg19.knownGene) txdb <- TxDb.Hsapiens.UCSC.hg19.knownGene keys <- head(keys(txdb, keytype="GENEID")) cols <- c("TXID", "TXSTART") select(txdb, keys=keys, columns=cols, keytype="GENEID")
Key Points
Bioconductor collates interoperable packages for analyses of biological data.
Using biocView can help you to find packages for a specific topic.